10 Basic Tidyverse Concepts

Figure 10.1: The Treachery of Images (Rene Magritte, 1948).
In this chapter we will introduce a few tools from the tidyverse set of R-packages:
- the pipe operator
%>%for chaining function calls in a convenient and readable way; - the
tibbleclass, a variant of the data frame that is especially suitable for large data sets; - data manipulation functions from the dplyr package suitable for use with the pipe operator:
-
filter()andselect()for sub-setting; -
mutate()for transforming variables; -
group_by()andsummarise()for numerical summaries of data.
-
10.1 The Tidyverse
The tidyverse isn’t a package, exactly—it’s a collection of packages. Go ahead and attach it:
You’ll get an account of the packages that have been attached. We have worked before with ggplot and by the end of CSC 215 we will have worked with all of the others. You need not worry about the fact that filter() and lag() mask functions from the stats package.
10.2 The magrittr Pipe Operator
In Section 6.8.4 you met R’s native pipe operator |>. The tidyverse uses another pipe operator, %>% from the magrittr package.26.
The keyboard shortcut for %>% is:
- Ctrl+Shift+M on Windows/Linux, or
- Cmd+Shift+M on Mac OS.
The the native R pipe operator, %>% connects two function calls by making the value returned by the first call the first argument of the second call. Here’s an example:
## [1] "hello" "hello" "hello" "hello"This is the same as the more familiar:
rep("hello", times = 4)## [1] "hello" "hello" "hello" "hello"Here’s another example:
# same as nrow(bcscr::m111survey)
bcscr::m111survey %>% nrow()## [1] 71Here’s two pipes:
## [1] 4By default the value of the left-hand call is piped into the right-hand call as the first argument. You can make it some other argument using the dot . as a placeholder, for example:
## [1] "hello" "hello" "hello" "hello"(Recall that the placeholder for the native R pipe is _. Do not interchange _ with .)
Since sub-setting is actually a function call under the hood, you can use the dot there, too:
## [1] 9The pipe operator isn’t all that useful when you only use it once or twice in succession. Its true value becomes apparent in the chaining together of many manipulations involving data frames.
10.2.1 Practice Exercises
-
Rewrite the following call with the tidyverse pipe operator, in three different ways:
seq(2, 22, by = 4)## [1] 2 6 10 14 18 22 -
Consider
mosaicData::CPS85:data("CPS85", package = "mosaicData")Use the pipe operator with
subset()to find the row ofmosaicData::CPS85containing the worker who made more than 40 dollars per hour. Display only the sex, age and wage of the worker.
10.3 Tibbles
The tibble package gives us tibbles, which are very nearly the same thing as a data frame. Indeed, the name “tibble” is supposed to remind us of a data “table.”
Consider the class of bcscr::m111survey:
class(bcscr::m111survey)## [1] "data.frame"Yep, it’s a data frame. But we can convert it to a tibble, as follows:
survey <- as_tibble(bcscr::m111survey)
class(survey)## [1] "tbl_df" "tbl" "data.frame"You can treat tibbles like data frames. For now the primary practical difference is manifest when you print a tibble to the Console:
survey## # A tibble: 71 × 12
## height ideal_ht sleep fastest weight_feel love_first extra_life seat GPA enough_Sleep
## <dbl> <dbl> <dbl> <int> <fct> <fct> <fct> <fct> <dbl> <fct>
## 1 76 78 9.5 119 1_underwei… no yes 1_fr… 3.56 no
## 2 74 76 7 110 2_about_ri… no yes 2_mi… 2.5 no
## 3 64 NA 9 85 2_about_ri… no no 2_mi… 3.8 no
## 4 62 65 7 100 1_underwei… no no 1_fr… 3.5 no
## 5 72 72 8 95 1_underwei… no yes 3_ba… 3.2 no
## 6 70.8 NA 10 100 3_overweig… no no 1_fr… 3.1 yes
## 7 70 72 4 85 2_about_ri… no yes 1_fr… 3.68 no
## 8 79 76 6 160 2_about_ri… no yes 3_ba… 2.7 yes
## 9 59 61 7 90 2_about_ri… no yes 3_ba… 2.8 no
## 10 67 67 7 90 3_overweig… no no 2_mi… NA yes
## # ℹ 61 more rows
## # ℹ 2 more variables: sex <fct>, diff.ideal.act. <dbl>The output is automatically truncated, and the number of columns printed is determined by the width of your screen. This is a great convenience when one is dealing with larger data sets.
Many larger data tables in packages will come to you as tibbles.
10.4 Subsetting with dplyr
The dplyr function filter() is the rough equivalent of select(): it picks out rows of a data frame (or similar objects such as a tibble). The dplyr function select() subsets for columns.
Thus you can use the two functions together to do perform sub-setting. With the pipe operator, your code can be quite easy to read:
survey %>%
filter((sex == "male" & height > 70) | (sex =="female" & height < 55)) %>%
select(sex, height, fastest)## # A tibble: 22 × 3
## sex height fastest
## <fct> <dbl> <int>
## 1 male 76 119
## 2 male 74 110
## 3 male 72 95
## 4 male 70.8 100
## 5 male 79 160
## 6 male 73 110
## 7 male 73 120
## 8 female 54 130
## 9 male 74 119
## 10 male 72 125
## # ℹ 12 more rowsNote that dplyr data-functions like filter() and select() take a data table as their first argument, and return a data table as well. Hence they may be chained together as we saw in the above example.
With select() it’s easy to leave out columns, too:
## # A tibble: 71 × 10
## height sleep fastest weight_feel extra_life seat GPA enough_Sleep sex
## <dbl> <dbl> <int> <fct> <fct> <fct> <dbl> <fct> <fct>
## 1 76 9.5 119 1_underweight yes 1_front 3.56 no male
## 2 74 7 110 2_about_right yes 2_middle 2.5 no male
## 3 64 9 85 2_about_right no 2_middle 3.8 no female
## 4 62 7 100 1_underweight no 1_front 3.5 no female
## 5 72 8 95 1_underweight yes 3_back 3.2 no male
## 6 70.8 10 100 3_overweight no 1_front 3.1 yes male
## 7 70 4 85 2_about_right yes 1_front 3.68 no male
## 8 79 6 160 2_about_right yes 3_back 2.7 yes male
## 9 59 7 90 2_about_right yes 3_back 2.8 no female
## 10 67 7 90 3_overweight no 2_middle NA yes female
## # ℹ 61 more rows
## # ℹ 1 more variable: diff.ideal.act. <dbl>10.4.1 Practice Exercises
Can you use the pipe to chain dplyr functions along with
nrow()to find out how many people insurveybelieve in love at first sight and drove more than 120 miles per hour?Find the three largest heights of the males who drove more than 120 miles per hour.
Use the pipe and
filter()to make violin plots of the wages of men and women inCPS85, where the outlier-person (whose wage was more than 40 dollars per hour) has been eliminated prior to making the graph.
10.5 Transforming Variables with dplyr
In dplyr you transform variables with the function mutate(). Here is an example:
## # A tibble: 71 × 3
## sex fastest dareDevil
## <fct> <int> <lgl>
## 1 male 119 FALSE
## 2 male 110 FALSE
## 3 female 85 FALSE
## 4 female 100 FALSE
## 5 male 95 FALSE
## 6 male 100 FALSE
## 7 male 85 FALSE
## 8 male 160 TRUE
## 9 female 90 FALSE
## 10 female 90 FALSE
## # ℹ 61 more rowsIn mutate() there is always a variable-name on the left-hand side of the = sign. It could be the same as an existing variable in the table if you are content to overwrite that variable. On the right side of the = is a function that can depend on variables in the data table.
You can transform more than one variable in a single call to mutate(), as in the code below. The output is shown in 10.2.
survey %>%
mutate(dareDevil = fastest > 125,
height_ft = height / 12) %>%
ggplot(aes(x = dareDevil, y = height_ft)) +
geom_boxplot(fill = "burlywood", out.alpha = 0) +
geom_jitter(width = 0.2) +
labs(
x = "Whether person drives more than 125 mph",
y = "height (ft)",
title = "Daredevils aren't any taller than cautious people!"
)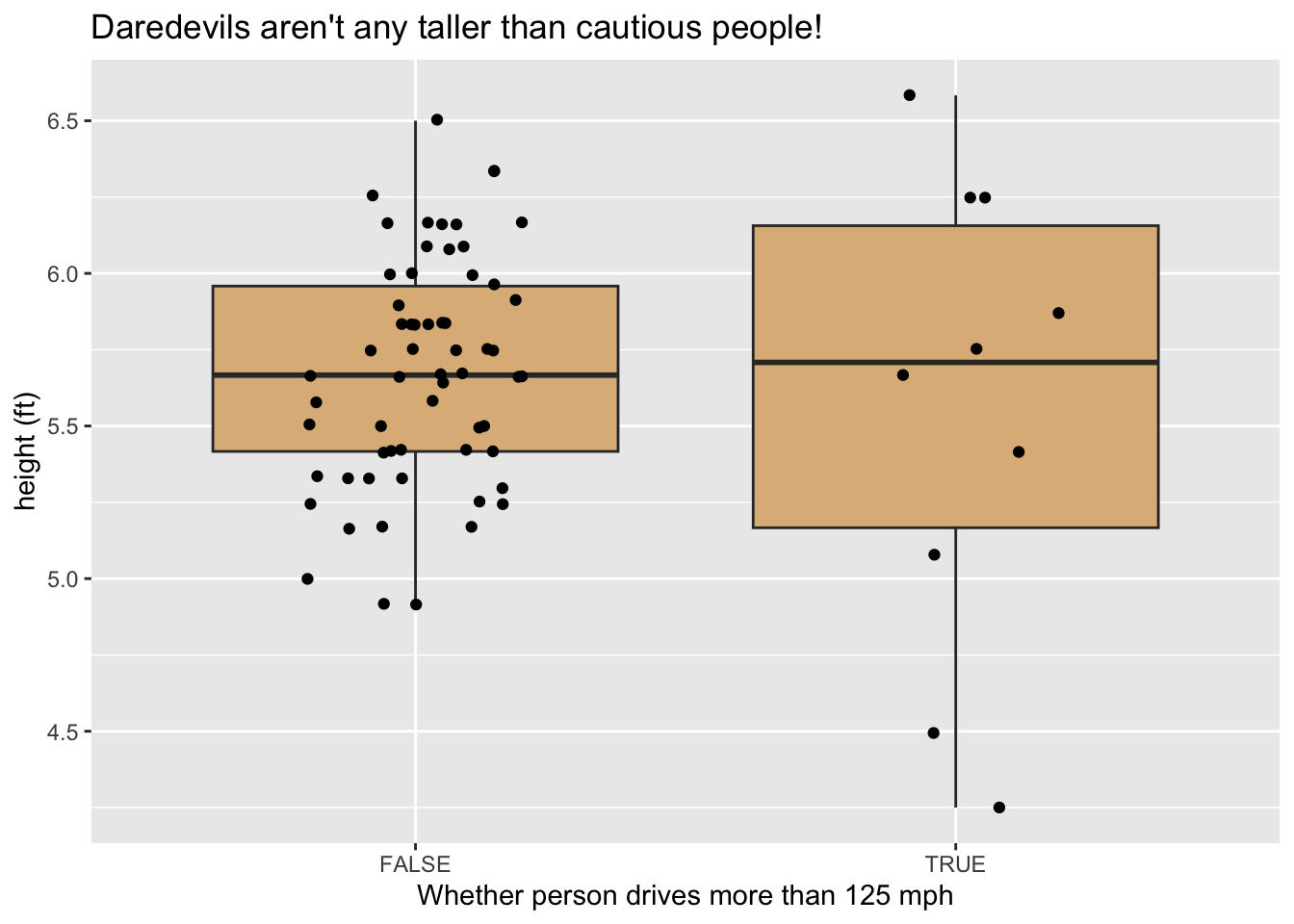
Figure 10.2: Graph produced after mutation.
10.5.1 Practice Exercises
- In
mosaicData::CPS85transform thewagevariable to units of dollars per day. (Assume an 8-hour working day.)
10.5.2 Solutions to Practice Exercises
-
Try this:
CPS85 %>% as_tibble() %>% # for display in Console mutate(dailyWage = wage * 8) %>% select(sex, sector, dailyWage) # for display in Console## # A tibble: 534 × 3 ## sex sector dailyWage ## <fct> <fct> <dbl> ## 1 M const 72 ## 2 M sales 44 ## 3 F sales 30.4 ## 4 F clerical 84 ## 5 M const 120 ## 6 F clerical 72 ## 7 F service 76.6 ## 8 M sales 120 ## 9 M manuf 88 ## 10 F sales 40 ## # ℹ 524 more rows
10.6 Grouping and Summaries
The next two dplyr data-functions are useful for generating numerical summaries of data.
Consider, for example, CPS85. We know from graphical studies that the men in the study are paid more than women, but how might we verify this fact numerically? One approach would be to separate the men and the women into two different groups and compute the mean wage for each group. This is accomplished by calling group_by() and summarise() in succession:
## # A tibble: 2 × 2
## sex meanWage
## <fct> <dbl>
## 1 F 7.88
## 2 M 9.99It’s possible to create more than one summary variable in a single call to summarise(), for example:
## # A tibble: 2 × 3
## sex meanWage n
## <fct> <dbl> <int>
## 1 F 7.88 245
## 2 M 9.99 289In the previous example, dplyr::n() was used to count the number of cases in each group.
For a more complete account of a numerical variable, one might consider the five-number summary:
- the minimum value
- the first quartile (Q1)
- the median
- the third quartile (Q3)
- the maximum value
These quantities are conveniently computed by R’s fivenum() function:
## [1] 1.00 5.25 7.78 11.25 44.50Let’s find the five number summaries for the wages of men and women:
CPS85 %>%
group_by(sex) %>%
summarise(
n = n(),
min = fivenum(wage)[1],
Q1 = fivenum(wage)[2],
median = fivenum(wage)[3],
Q3 = fivenum(wage)[4],
max = fivenum(wage)[5]
)## # A tibble: 2 × 7
## sex n min Q1 median Q3 max
## <fct> <int> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 F 245 1.75 4.75 6.8 10 44.5
## 2 M 289 1 6 8.93 13 26.3It’s also possible to group by more than one variable at a time. For example, suppose that we wish to compare the wages of men and women in the various sectors of employment. All we need to do is group by both sex and sector:
CPS85 %>%
group_by(sector, sex) %>%
summarise(
n = n(),
min = fivenum(wage)[1],
Q1 = fivenum(wage)[2],
median = fivenum(wage)[3],
Q3 = fivenum(wage)[4],
max = fivenum(wage)[5]
)## # A tibble: 15 × 8
## # Groups: sector [8]
## sector sex n min Q1 median Q3 max
## <fct> <fct> <int> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 clerical F 76 3 5.1 7 9.55 15.0
## 2 clerical M 21 3.35 6 7.69 9 12
## 3 const M 20 3.75 7.15 9.75 11.8 15
## 4 manag F 21 3.64 6.88 10 11.2 44.5
## 5 manag M 34 1 8.8 14.0 18.2 26.3
## 6 manuf F 24 3 4.36 4.9 6.05 18.5
## 7 manuf M 44 3.35 6.58 8.94 11.2 22.2
## 8 other F 6 3.75 4 5.62 6.88 8.93
## 9 other M 62 2.85 5.25 7.5 11.2 26
## 10 prof F 52 4.35 7.02 10 12.3 25.0
## 11 prof M 53 5 8 12 16.4 25.0
## 12 sales F 17 3.35 3.8 4.55 5.65 14.3
## 13 sales M 21 3.5 5.56 9.42 12.5 20.0
## 14 service F 49 1.75 3.75 5 8 13.1
## 15 service M 34 2.01 4.15 5.89 8.75 25Note that there were no women in the construction sector, so that group did not appear in the summary.
10.6.1 Note on Binding
Keep in mind that you can always “save” the results of any computation by binding them to a variable name, thus:
sexSector <-
CPS85 %>%
group_by(sector, sex) %>%
summarise(
n = n(),
min = fivenum(wage)[1],
Q1 = fivenum(wage)[2],
median = fivenum(wage)[3],
Q3 = fivenum(wage)[4],
max = fivenum(wage)[5]
)
class(sexSector)## [1] "grouped_df" "tbl_df" "tbl" "data.frame"Note that the result has data.frame as one of its classes, so you may extract components in any of the ways you have learned. The old ways, for instance, are fine:
# minimum wage among male professionals:
with(sexSector, min[sex == "M" & sector == "prof"])## [1] 510.6.2 Practice Exercises
These exercises deal with flight data from the nycflights13 data frame:
data("flights", package = "nycflights13")The
flightstable gives information about each departure in the year 2013 from one of the three major airports near New York City: John F. Kennedy (JFK), LaGuardia (LGA) or Newark (EWR). The airport from which the plane departed is recorded in the variableorigin. The variabledep_delaygives the delay in departure, in minutes. (This is a negative number if the plane left early). Find the number of departures and the mean departure delay for each of the three airports. (Note thatdep_delayfor cancelled flights will beNA.)The variable
distancegives the distance, in miles, between an origin and destination airport. For July 26, 2013, make a violin plot of the distances traveled by the departing planes from the each of the three New York airports. Use the pipe andfilter()to takeflightsinto the desired plot.Examine the plot you made in the previous problem: two of the flights appear to be about 5000 miles. Use the pipe,
filter()andselect()to display the origin, destination and distance for these two flights.
10.6.3 Solutions to Practice Exercises
-
Flights that were cancelled have
NAfor their departure delay, so we need to filter out these cases first, in order to correctly count the number of flights that actually left the airport. Try this:flights %>% filter(!is.na(dep_delay)) %>% group_by(origin) %>% summarise( departures = n(), meanDelay = mean(dep_delay) )## # A tibble: 3 × 3 ## origin departures meanDelay ## <chr> <int> <dbl> ## 1 EWR 117596 15.1 ## 2 JFK 109416 12.1 ## 3 LGA 101509 10.3 -
Try this:
flights %>% filter(month == 6 & day == 26) %>% ggplot(aes(x = origin, y = distance)) + geom_violin(fill = "burlywood") + geom_jitter(width = 0.25, size = 0.1) -
Try this:
## # A tibble: 2 × 3 ## origin dest distance ## <chr> <chr> <dbl> ## 1 JFK HNL 4983 ## 2 EWR HNL 4963
10.7 R’s Native Pipe
The magritter pipe became so popular that the R Core Team decided to include a pipe operator in R’s base package. It looks like this: |>.
For our purposes, R’s “native” pipe can be used interchangeably with %>%, except that the placeholder is indicated by the underscore (_) instead of a period.
A few examples:
letters |>
## letters is passed as the first argument of rep:
rep(times = 2)## [1] "a" "b" "c" "d" "e" "f" "g" "h" "i" "j" "k" "l" "m" "n" "o" "p" "q" "r" "s" "t" "u"
## [22] "v" "w" "x" "y" "z" "a" "b" "c" "d" "e" "f" "g" "h" "i" "j" "k" "l" "m" "n" "o" "p"
## [43] "q" "r" "s" "t" "u" "v" "w" "x" "y" "z"
letters |>
## extract the first five elements of letters:
_[1:5] |>
#3 pass these as the first argument of rep:
rep(times = 1:5)## [1] "a" "b" "b" "c" "c" "c" "d" "d" "d" "d" "e" "e" "e" "e" "e"
1:length(letters) |>
## the placeholder below passes the above vector
## to the second argument of rep:
rep(letters, times = _)## [1] "a" "b" "b" "c" "c" "c" "d" "d" "d" "d" "e" "e" "e" "e" "e" "f" "f" "f" "f" "f" "f"
## [22] "g" "g" "g" "g" "g" "g" "g" "h" "h" "h" "h" "h" "h" "h" "h" "i" "i" "i" "i" "i" "i"
## [43] "i" "i" "i" "j" "j" "j" "j" "j" "j" "j" "j" "j" "j" "k" "k" "k" "k" "k" "k" "k" "k"
## [64] "k" "k" "k" "l" "l" "l" "l" "l" "l" "l" "l" "l" "l" "l" "l" "m" "m" "m" "m" "m" "m"
## [85] "m" "m" "m" "m" "m" "m" "m" "n" "n" "n" "n" "n" "n" "n" "n" "n" "n" "n" "n" "n" "n"
## [106] "o" "o" "o" "o" "o" "o" "o" "o" "o" "o" "o" "o" "o" "o" "o" "p" "p" "p" "p" "p" "p"
## [127] "p" "p" "p" "p" "p" "p" "p" "p" "p" "p" "q" "q" "q" "q" "q" "q" "q" "q" "q" "q" "q"
## [148] "q" "q" "q" "q" "q" "q" "r" "r" "r" "r" "r" "r" "r" "r" "r" "r" "r" "r" "r" "r" "r"
## [169] "r" "r" "r" "s" "s" "s" "s" "s" "s" "s" "s" "s" "s" "s" "s" "s" "s" "s" "s" "s" "s"
## [190] "s" "t" "t" "t" "t" "t" "t" "t" "t" "t" "t" "t" "t" "t" "t" "t" "t" "t" "t" "t" "t"
## [211] "u" "u" "u" "u" "u" "u" "u" "u" "u" "u" "u" "u" "u" "u" "u" "u" "u" "u" "u" "u" "u"
## [232] "v" "v" "v" "v" "v" "v" "v" "v" "v" "v" "v" "v" "v" "v" "v" "v" "v" "v" "v" "v" "v"
## [253] "v" "w" "w" "w" "w" "w" "w" "w" "w" "w" "w" "w" "w" "w" "w" "w" "w" "w" "w" "w" "w"
## [274] "w" "w" "w" "x" "x" "x" "x" "x" "x" "x" "x" "x" "x" "x" "x" "x" "x" "x" "x" "x" "x"
## [295] "x" "x" "x" "x" "x" "x" "y" "y" "y" "y" "y" "y" "y" "y" "y" "y" "y" "y" "y" "y" "y"
## [316] "y" "y" "y" "y" "y" "y" "y" "y" "y" "y" "z" "z" "z" "z" "z" "z" "z" "z" "z" "z" "z"
## [337] "z" "z" "z" "z" "z" "z" "z" "z" "z" "z" "z" "z" "z" "z" "z"Exercises

-
Use the pipe operator to rewrite the following command in three ways:
runif(10, min = 0, max = 5) -
Rewrite the following command using two pipe operators in succession:
paste("hello", "there", "Bella")## [1] "hello there Bella" -
Use the pipe operator and dplyr functions to rewrite the following command:
The next few exercises are about the
babynamesdata frame from the babynames package. Find the names for females born in 2015 that were given to more than 1% of female applicants (i.e.,
propis bigger than 0.01).-
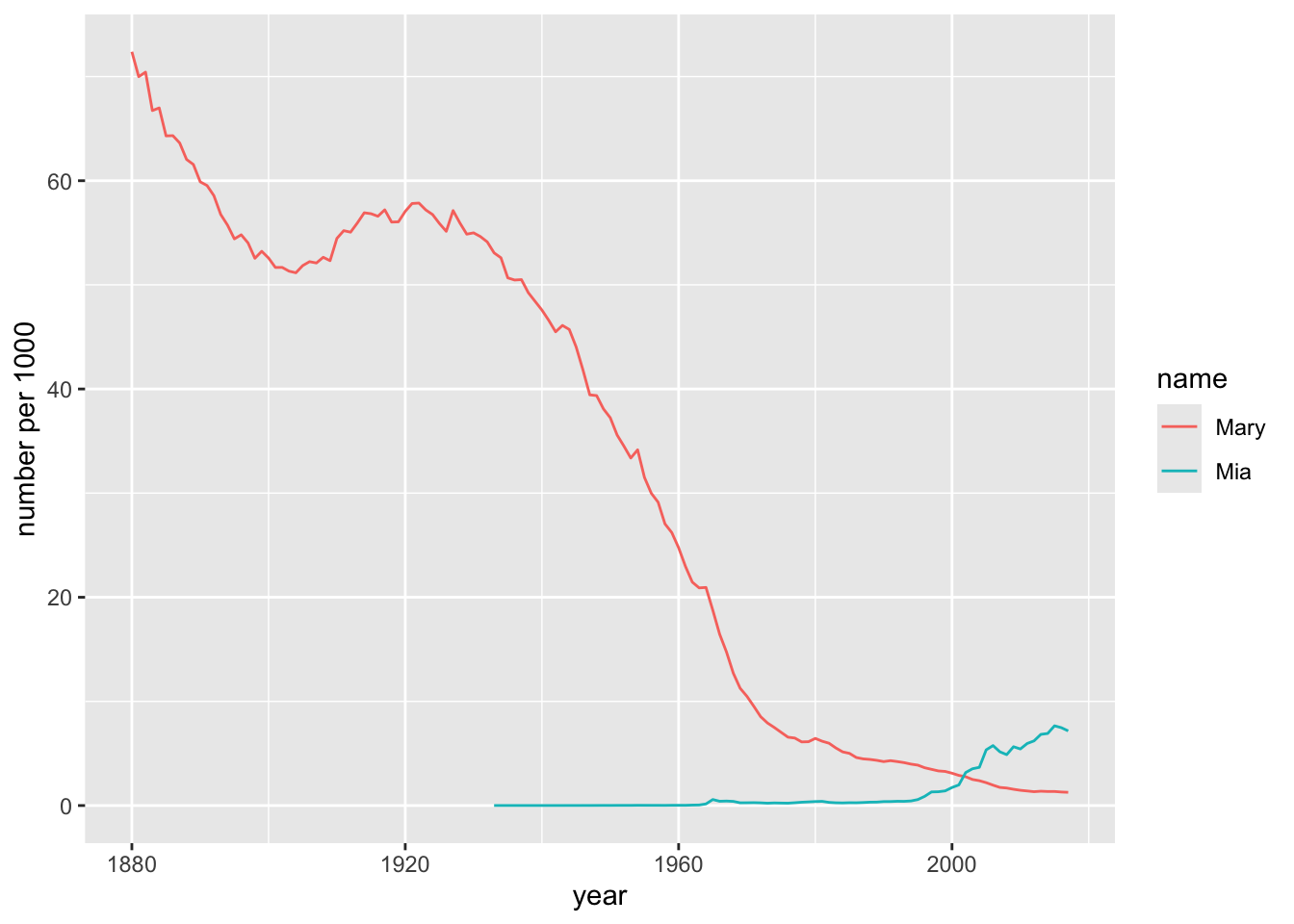
Use the pipe operator and dplyr functions to produce the following graph of the popularity of “Mary” and “Mia” as girl-names over the years. Note that popularity is given as number per one thousand applicants, i.e., as
prop * 1000.